The discovery of a key DNA sequence adds weight to a theory on the costs of reproduction.
A theory that some genes provide benefits in early life — allowing organisms to have more offspring — but detrimental effects in later life, leading to aging, has been given a boost by Tsinghua University researchers who have found one such a gene.

Two Caenorhabditis elegans nematode worms 60 mins after they have been fed after being starved. The top worm is a wild type and shows less translation of proteins associated with egg yolks than the bottom, in which the trl-1 gene is deleted.
This finding in a tiny nematode worm lends weight to the so-called ‘antagonistic pleiotropy’ theory of aging, which was proposed in 1957 and posits that there is a trade-off between early reproductive fitness and lifespan1. The results have been published in the journal PNAS2.
Under this theory, aging is driven by pleiotropic genes — these influence two or more seemingly unrelated physical traits which offer a reproductively useful advantage early in life, but later become detrimental. But until now the theory has lacked direct genetic evidence.
“We think antagonistic pleiotropy is either very common or ubiquitous throughout the animal world,” says lead researcher behind the study, Professor Guangshuo Ou. Quantitative genetic studies and studies of naturally segregating polymorphisms, have hinted at the trade-off between traits, but other explanations couldn’t be ruled, he explains. “Compelling evidence of antagonistic pleiotropy at an individual gene resolution remains scarce.”
The researchers made their breakthrough by studying a nematode worm called Caenorhabditis elegans, which is commonly used as a model organism.
C. elegans adapts rapidly when under environmental stress to enhance its chance of survival and ensure growth and reproduction. For instance, in response to an absence of food during development, worms turns into a potentially long-lived ‘dauer larvae’ incapable of reproduction. But on refeeding, they revert to their former state.
By studying the biochemistry of worms recovering from starvation, Ou’s team have identified a likely antagonistic pleiotropic gene that directs nutrients to egg development within well-fed worms.
Feeding offspring ages parents
By performing genetic analyses of C. elegans following six days of starvation, they found that a gene dubbed trl-1 was producing more proteins after refeeding. To test if trl-1 was linked to reproduction and lifespan, they created a strain of C. elegans devoid of the gene, and found that they “had increased brood sizes and shortened lifespans,” Ou says.
Furthermore, his team also showed that genes which help produce a yolk protein, called vitellogenin, were upregulated in the modified worms.
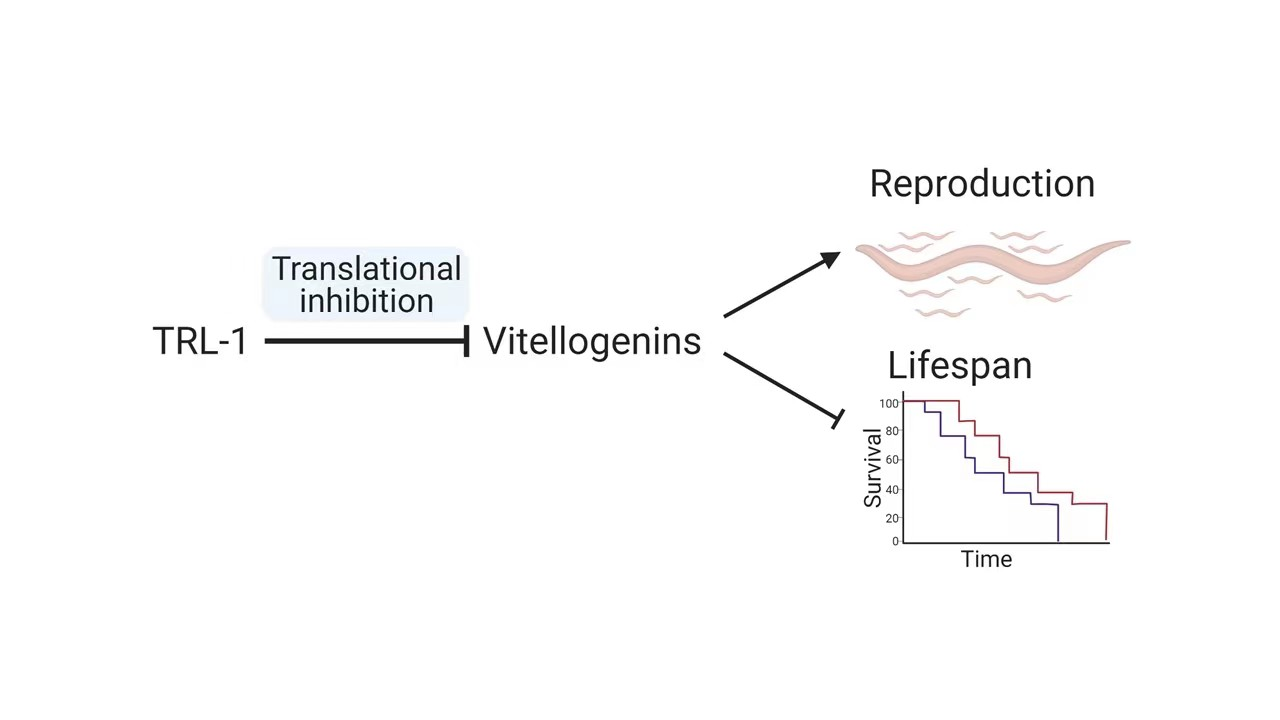
The TRL-1 protein inhibits vitellogenin messenger RNA translation, and thus it reduces the production of egg yolk proteins. However, this also extends the lifespan of the parent nematode.
“The loss of trl-1 upregulated the translation of vitellogenin, which produces copious yolk to provision eggs,” Ou explains. But the overexpression of vitellogenin also “is also known to reduce the lifespan in C. elegans3,” he adds.
The researchers then went on to show that the protein produced by trl-1 works to inhibit the translation of vitellogenin messenger RNA to halt the production of the egg yolk protein.
The end result is that nutrients provided to developing eggs are limited, but the lifespan of the parent nematode is extended because it is putting fewer resources into egg development.
“These results indicate that trl-1 functions as an antagonistic pleiotropic gene that regulates the reproduction-longevity trade-off by optimizing nutrient production for eggs,” says Ou.
He adds that while analyses had not previously identified similar gene sequences in mammals, “functional gene sequences, rather than protein sequences, are more likely to be preserved”.
The team is now searching for other genes involved in recovery from starvation, which might also be linked to a trade-off in reproductive fitness and lifespan.
“Understanding trl-1 has already helped to clarify one aspect of how the genome shapes the aging process, and this will help us study of the genetic factors that influence longevity and age-associated diseases,” he says.
References
1. Williams, G. C. Pleiotropy, Natural Selection, and the Evolution of Senescence. Evolution, 11, 398-411 (1957).
2. Wu, D., Wang, Z., Huang, J., Huang, L., Zhang ,S. et al. An antagonistic pleiotropic gene regulates the reproduction and longevity tradeoff. Proceedings of the Notational Academy of Science U S A. 119(18), e2120311119 (2022). doi: 10.1073/pnas.2120311119
3. Seah, N., E., de Magalhaes Filho, C., D., Petrashen, A., P., Henderson, H., R., Laguer, J., et al. Autophagy-mediated longevity is modulated by lipoprotein biogenesis Autophagy 12(2), 261–272 (2016) doi: 10.1080/15548627.2015.1127464
Editor: Guo Lili