Long non-coding RNAs (lncRNAs) have emerged as important regulators in various physiological processes and disease, including development and tumorigenesis1-3. Despite their significance, the functions of most lncRNAs remain unknown. Homologous lncRNAs that are conserved across different species are more likely to possess important functions. However, due to the low sequence conservation of lncRNAs4,5, traditional sequence alignment methods can only identify a limited number of homologous lncRNAs between species. For instance, there are tens of thousands of lncRNAs in human and zebrafish, only a few dozen homologous lncRNAs can be identified through sequence alignment. There is, therefore, an urgent need for a new method to identify homologous lncRNAs.
On January 9, 2024, a research paper by Qiangfeng Zhang’s group from Tsinghua University, Yangming Wang’s group and Jianzhong Xi’s group from Peking University was published in Nature Genetics. This study, titled "Computational prediction and experimental validation identify functionally conserved lncRNAs from zebrafish to human" presents a novel computational method for identifying homologous lncRNAs across eight vertebrate species, including humans, mice, and zebrafish. They also developed a CRISPR-based knockout screen and rescue system to experimentally validate the conserved function of the predicted homologous lncRNAs in different species, thereby providing new insights for future research in this field.
This study introduces a computational method (lncHOME), which integrates genome comparison and machine learning to identify lncRNAs with conserved genomic positions and patterns of RNA binding protein (RBP) binding sites across eight vertebrate species; these lncRNAs were defined as coPARSE-lncRNAs (Figure 1).
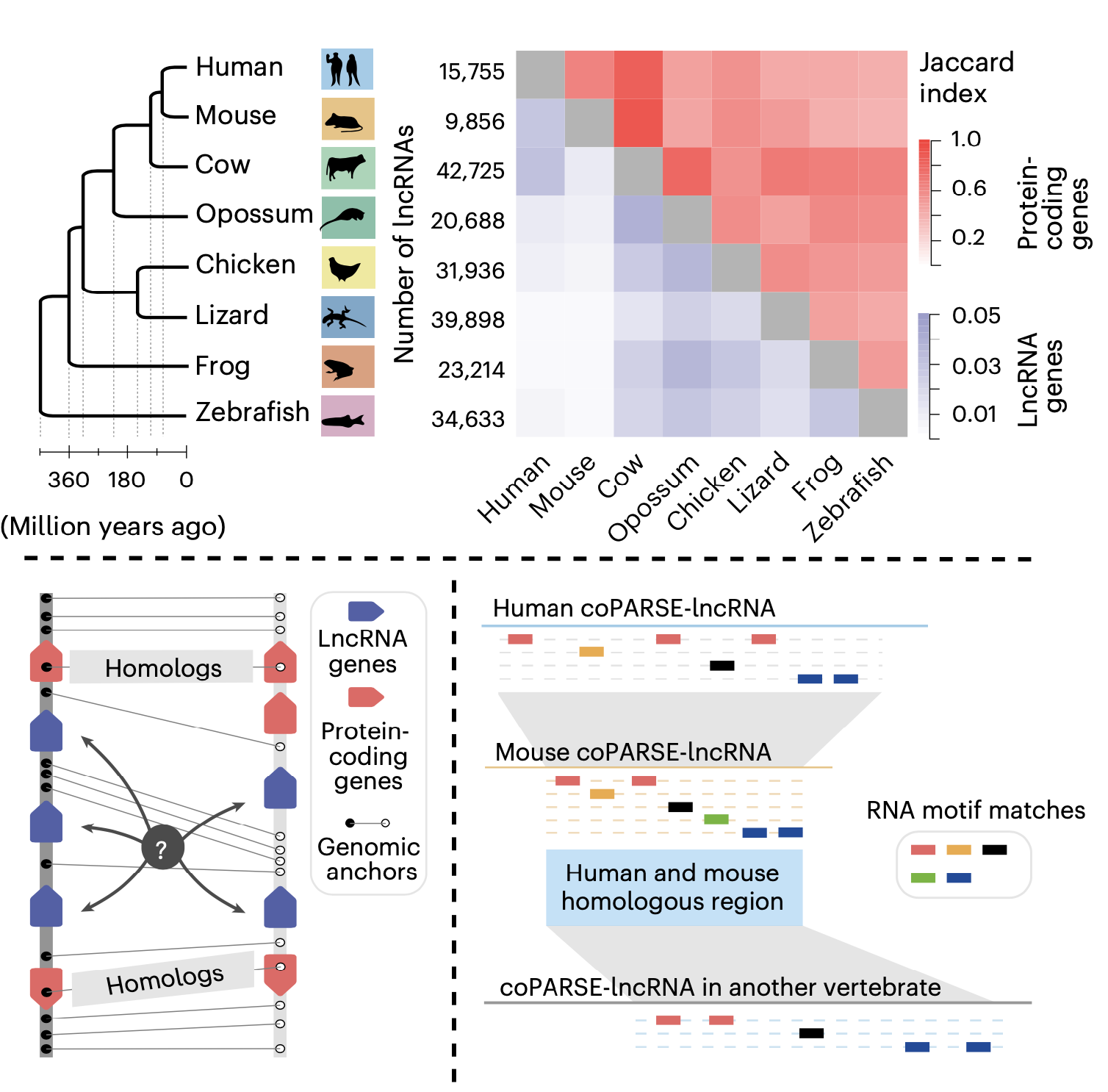
Figure 1. The computational method (lncHOME) for homologous lncRNA identification
lncHOME identified 570 human coPARSE-lncRNAs with predicted homologs in zebrafish, of which only 17 homologous lncRNA pairs could be identified through sequence alignment. Compared to non-homologous lncRNAs, these coPARSE-lncRNAs are enriched for disease-associated mutations and are more likely to be dysregulated in cancer tissues, suggesting that these coPARSE-lncRNAs may have important physiological functions.
To functionally characterize these coPARSE-lncRNAs, the study developed a CRISPR-Cas12a knockout screen and identified 75 coPARSE-lncRNAs that promote cell proliferation. The researchers further developed a single-step knockout-rescue approach, also based on CRISPR-Cas12a, to assess the functional conservation of lncRNA homolog pairs. They discovered that the knockout of four human coPARSE-lncRNAs resulted in defects in HeLa cell proliferation, which were subsequently rescued by the predicted zebrafish homologs. Intriguingly, knocking down these four zebrafish coPARSE-lncRNAs in zebrafish embryos caused severe developmental delays, which were rescued by the predicted human homologs. These results strongly support the functional conservation of these predicted homologous lncRNAs.
The homologous lncRNAs identified by the lncHOME exhibit conserved patterns of RBP binding sites. Based on this observation, it is speculated that the coPARSE-lncRNAs and their predicted homologs share similar RBP binding profiles. To validate this hypothesis, this study performed RNA pull-down followed by mass spectrometry experiments on two coPARSE-lncRNAs and their predicted homologs. The results confirmed that these lncRNAs indeed exhibited similar RBP binding profiles.
Furthermore, the study investigated the functional significance of predicted RBP binding sites in the conserved function of coPARSE-lncRNAs. It introduced mutations into the binding sites of specific RBPs (such as NONO and IGF2BP2) in the aforementioned homologous lncRNA fragments that were capable of rescuing cell proliferation or embryonic developmental defects. The experiments showed that the mutations decreased the rescue effects of the lncRNA fragments. These mutation experiments demonstrate the importance of RBP binding sites for the conserved function of coPARSE-lncRNAs.
In summary, this study provides a machine learning-based computational method that identifies a series of potential homologous lncRNAs in vertebrates. They experimentally validated the functional conservation of homologous lncRNAs. Although the sequence conservation of these lncRNAs diminishes during evolution, the conserved RBP binding patterns have been retained (Figure 2). This work greatly expands the current repository of conserved lncRNAs in vertebrates and provides new perspectives and resources for studying the evolution, function, and mechanisms of lncRNAs.
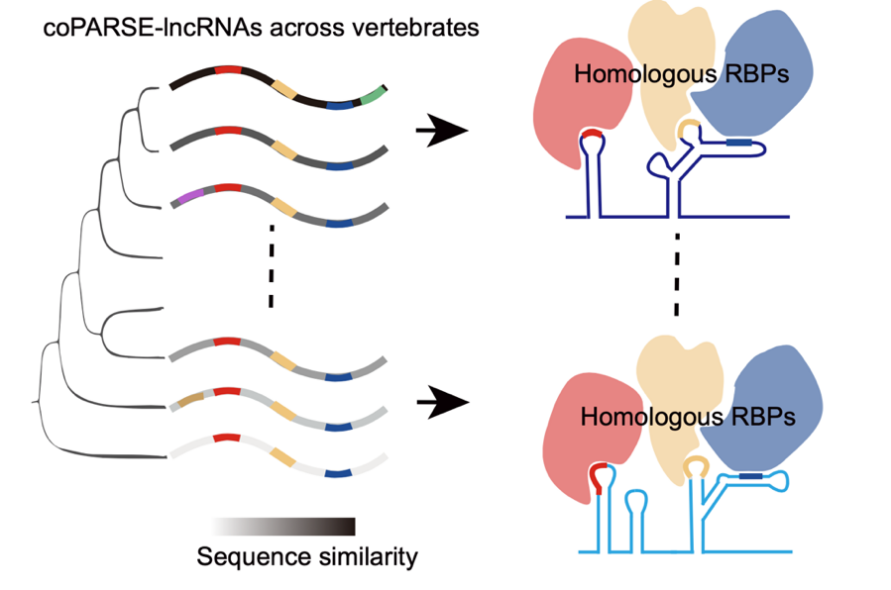
Figure 2. The model for the evolution and function of coPARSE-lncRNAs.
Associate Professor Qiangfeng Zhang from the School of Life Sciences, Tsinghua University, and Professor Yangming Wang and Professor Jianzhong Xi from the Future Technology Institute, Peking University, are the corresponding authors of the paper. Dr. Wenze Huang and Dr. Tuanlin Xiong from the School of Life Sciences, Tsinghua University, along with Dr. Yuting Zhao from the Future Technology Institute, Peking University, are the co-first authors of the paper. Professor Feng Liu and Dr. Jian Heng from Institute of Zoology, Chinese Academy of Sciences, Dr. Ge Han and Pengfei Wang from the School of Life Sciences, Tsinghua University, Dr. Zhihua Zhao, Dr. Juan Li, Ming Shi, Jiazhen Wang, and Yixia Wu have made significant contributions to the research work.
Link: https://www.nature.com/articles/s41588-023-01620-7
Editor: Li Han

