Xianyang Fang's group and Cheng-feng Qin’s group published a research article in EMBO Reports revealing extended and flexible 3D structures for the long non-coding subgenomic flavivirus RNAs in solution
On September 10th 2019, Xianyang Fang’s group from the School of Life Sciences of Tsinghua University and Cheng-feng Qin’s group from the Beijing Institute of Microbiology and Epidemiology published an article entitled “Long non-coding subgenomic flavivirus RNAs have extended 3D structures and are flexible in solution” in EMBO Reports. For the first time, it was found that the long non-coding subgenomic RNAs from type 2 dengue virus, Zika virus and West Nile virus have extended 3D structures and are flexible in solution.
The field of long non-coding RNAs (lncRNA) is a new frontier in life sciences. lncRNAs are transcripts that have no protein-coding potential but are larger than 200 nts in length and play diverse roles in many biological processes. Many if not all mosquito-borne flaviviruses, such as dengue virus (DENV), Zika virus (ZIKV) and West Nile virus (WNV), produce a large abundance of long non-coding subgenomic RNAs (sfRNAs) in infected cells that link to viral pathogenicity and host immune evasion. Until now, the structural characterization of these lncRNAs remain limited, as structural studies of RNAs using traditional biophysical techniques, including X-ray crystallography, NMR, and single-particle cryo-electron microscopy are extremely challenging. In this study, Xianyang Fang’s group and collaborators studied the 3D structures of individual and combined subdomains of sfRNAs and visualized the accessible 3D conformational spaces of complete sfRNAs from DENV2, ZIKV and WNV, using small angle x-ray scattering (SAXS) and computational modeling. The study found that the long non-coding subgenomic RNAs consist of multiple sub-domains, which together are organized into extended three-dimensional structures with high flexibility in solution, resembling the modular structural features in multi-domain proteins. It is interesting to note that the 3D structures of the duplicated dumbbell (DB12s) sub-domains of dengue virus, Zika virus and West Nile virus are significantly different, which can be explained by the differences in the topology of pseudoknots arrangement in DB12s, and confirmed by in vitro biochemical and in vivo cell biology experiments. The work not only provides structural insight into the function of flavivirus sfRNAs, but also highlights the strategies of visualizing the 3D structures of other lncRNAs in solution by SAXS and computational methods (Figure 1).
Yupeng Zhang (Ph.D. student of the School of Life Sciences in Tsinghua University), Yikan Zhang (Ph.D. student of the School of Life Sciences in Tsinghua University), Zhong-Yu Liu (Associate professor at the School of Medicine (Shenzhen), Sun Yat-sen University) and Meng-Li Cheng (Graduate student of the Beijing Institute of Microbiology and Epidemiology) are co-first authors of this article. Both Junfeng Ma and Yan Wang from Xianyang Fang’s group contributed to this study. Dr. Xianyang Fang from Tsinghua University and Dr. Cheng-feng Qin from the Beijing Institute of Microbiology and Epidemiology are co-corresponding authors of this article. This study was supported by grants from the National Key Research and Development Project of China, Tsinghua University Initiative Scientific Research Program, the Beijing Advanced Innovation Center for Structural Biology and the Tsinghua-Peking Joint Center for Life Sciences. The SAXS data was collected at the beamline 12-ID-B, Advanced Photon Source, Argonne National Laboratory, USA and the beamline BL19U2, Shanghai Synchrotron Radiation Facility, China.
A link to the paper can be found here:https://doi.org/10.15252/embr.201847016
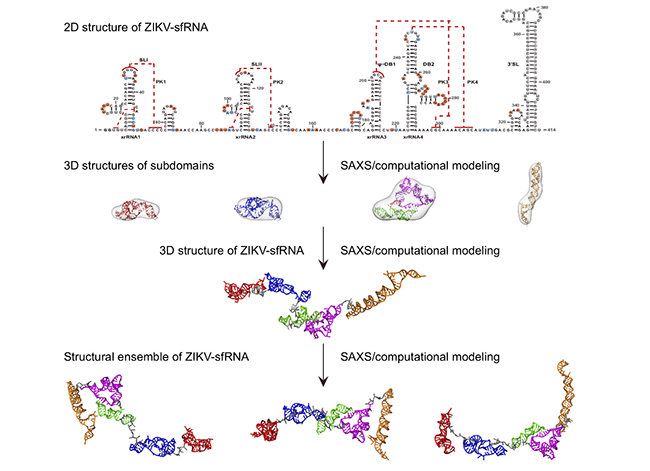
Figure 1: A General protocol for 3D structural study of the long non-coding subgenomic flavivirus RNA from ZIKV by SAXS and computational modeling.
From School of Life Sciences

