Xuerui Yang’s group developed an integrative multi-omics data-mining strategy to dissect the cancer type-specific transcriptional regulation circuitry dependent on the promoter DNA methylome
Transcription regulatory networks (TRNs), which are composed of regulatory circuits from TFs to their target genes, provide detailed maps of gene expression regulations in specific cellular contexts. Despite the extensive findings about interplays between DNA methylation and TFs in determining the gene expression profile, comprehensive understandings about how the DNA methylome is incorporated in the cancer-specific transcriptional regulation circuitry and involved in regulating the gene expression abnormality in cancers have been missing. Dynamic dysregulation of the promoter DNA methylome is a signature of cancer. However, a comprehensive survey of the potential functions of the cancer context-specific DNA methylome, which should have substantial value for understanding the multi-level gene expression regulation machinery in cancers, has never been done at the single-CpG site level.
In a recently published paper by Cell Reports, Dr. Xuerui Yang’s group introduced an integrative analysis pipeline based on mutual information theory, to systematically find dependencies of transcriptional regulation circuits on promoter CpG methylation profiles. By taking advantages of the multi-omics tumor profiling data in The Cancer Genome Atlas (TCGA), their analyses generated a series of Cancer type-specific DNA Methylation-dependent Transcription Regulatory Networks (MeTRNs), for 21 major cancer types.
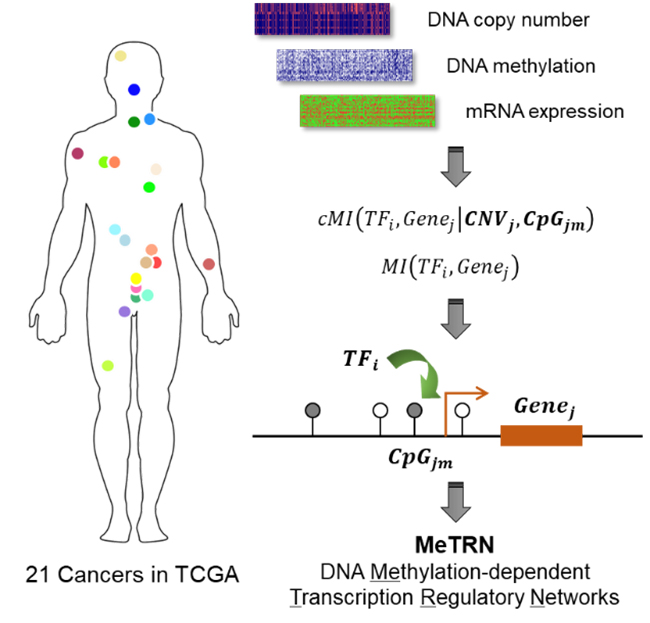
Scheme of the multi-omics data analysis strategy
By coupling specific transcription factors with CpG sites, such cancer type-specific transcriptional regulation circuitry serves as an indispensable layer of expression regulation for many key cancer-related genes. The coupled CpG sites and transcription factors also serve as markers for classifications of cancer subtypes with different prognoses, suggesting physiological relevance of the regulation machinery. Therefore, the value of the work lies in its multi-omics data-mining methodology and its potential as a new framework and resource for further studies of the epigenetic scheme in gene expression dysregulations in cancers.
Yu Liu, Yang Liu, and Rongyao Huang in Xuerui Yang’s lab performed most of the analyses. The study was financially supported by the national key research and development program, Precision Medicine Project and the National Natural Science Foundation of China. The study also received strong supports from the High-Performance Computing platform of the National Protein Science Facility (Beijing), at Tsinghua University.
Link to the paper: https://www.cell.com/cell-reports/fulltext/S2211-1247(19)30270-0
(From School of Life Sciences)

