Professor Yijun Qi’s team from the School of Life Sciences at Tsinghua University recently published significant findings on the modes of action and roles of maize phased small interfering RNAs (phasiRNAs). Using low-input degradome-sequencing, they analyzed maize anthers and isolated male germ cells at various developmental stages. Their study identified hundreds of protein-coding mRNAs as 21-nt phasiRNA targets and revealed that 21-nt phasiRNA-directed mRNA cleavage is temporally dynamic and cell type-dependent. Notably, while 21-nt phasiRNAs can direct target mRNA cleavage and regulate male fertility in both maize and rice, the sequences of 21-nt phasiRNAs and their targets show little conservation in these two species.
Twenty-one- and 24-nt (phasiRNAs) are two classes of small RNAs abundantly expressed in developing grass anthers. They play crucial roles in anther development and male reproduction, as dysfunction of these phasiRNAs can lead to photo/thermo-sensitive male sterility. Previous studies have revealed that 21-nt phasiRNAs in rice direct the cleavage of target mRNAs to regulate gene expression. However, it remains elusive whether 21-nt phasiRNAs target mRNAs in maize.
To experimentally test whether 21-nt phasiRNAs can direct mRNA cleavage, and to determine in which cell types they act in maize, Professor Qi’s team collected anthers at premeiotic, prophase I and tetrad/microspore stages as well as highly pure meiocytes at prophase I, tetrads, and microspores from wild-type maize (Fig. 1). Through low-input small RNA-sequencing, they identified 10,314 21-nt phasiRNAs from these samples. They then applied low-input degradome-sequencing to detect 21-nt phasiRNA-directed cleavage signals. Data analysis revealed that 21-nt phasiRNAs directed the cleavage of 132, 132 and 119 mRNAs in premeiotic, prophase I and tetrad/microspore-stage anthers and they directed the cleavage of 246, 181 and 143 mRNAs in prophase I meiocytes, tetrads and microspores, respectively. The results suggest that 21-nt phasiRNAs can direct mRNA cleavage in maize anthers and male germ cells.
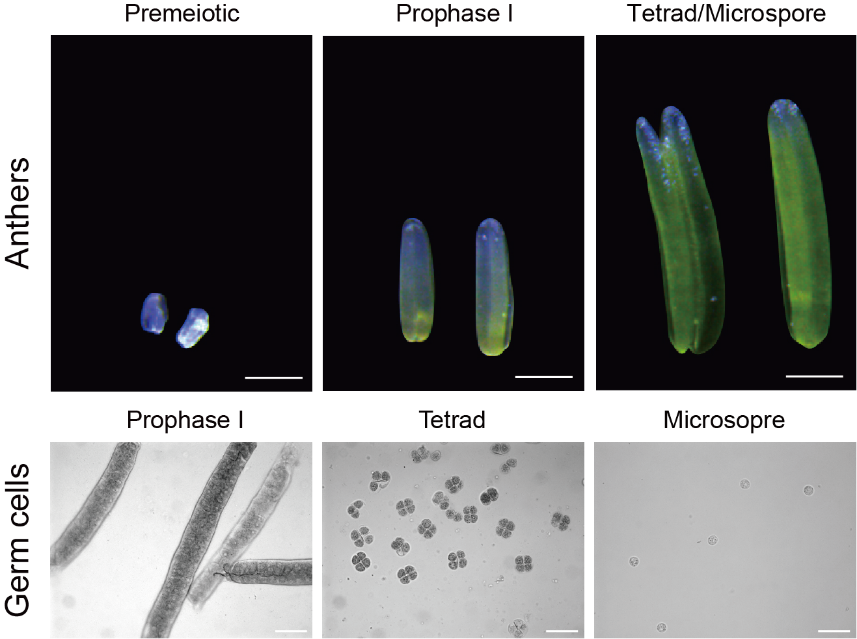
Fig. 1. Maize anthers and male germ cells at various developmental stages
They found that 21-nt phasiRNA-directed cleavage signals were dynamic across various stages of anther and male germ cell development (Fig. 2), suggesting that 21-nt phasiRNAs direct target mRNA cleavage in a temporally specific manner. In anthers and male germ cells at the same developmental stage, some 21-nt phasiRNAs targeted genes specifically expressed in the somatic anther wall cells or male germ cells (Fig. 3). These target genes are associated with specific biological functions of the somatic anther wall cells and male germ cells, respectively, suggesting that 21-nt phasiRNAs direct target mRNA cleavage in a cell type-dependent manner. Professor Qi’s team previously found that 5'-terminal nucleotide of small RNAs determines their binding specificity to effector AGO proteins. They found that compared to 21-nt phasiRNAs that do not have any targets, those with targets only in male germ cells exhibited a very strong preference for C at their 5′ ends. The preference was less evident for 21-nt phasiRNAs with targets in both anthers and male germ cells, and it was further attenuated for those with targets exclusively in anthers. These results indicate that distinct populations of 21-nt phasiRNAs exist in somatic anther wall cells and male germ cells, which target unique sets of mRNAs through binding to different AGO effector proteins.
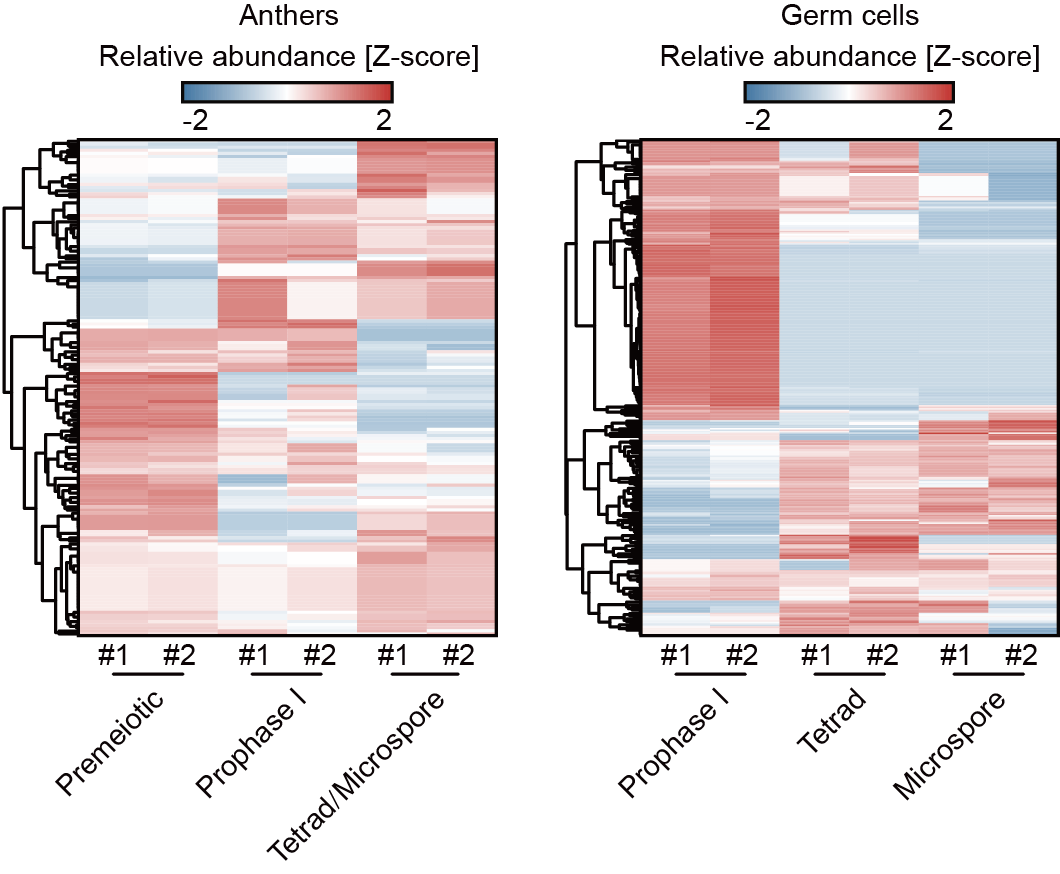
Fig. 2 Twenty-one-nt phasiRNA-directed cleavage signals at premeiotic, prophase I, tetrad and microspore stages.
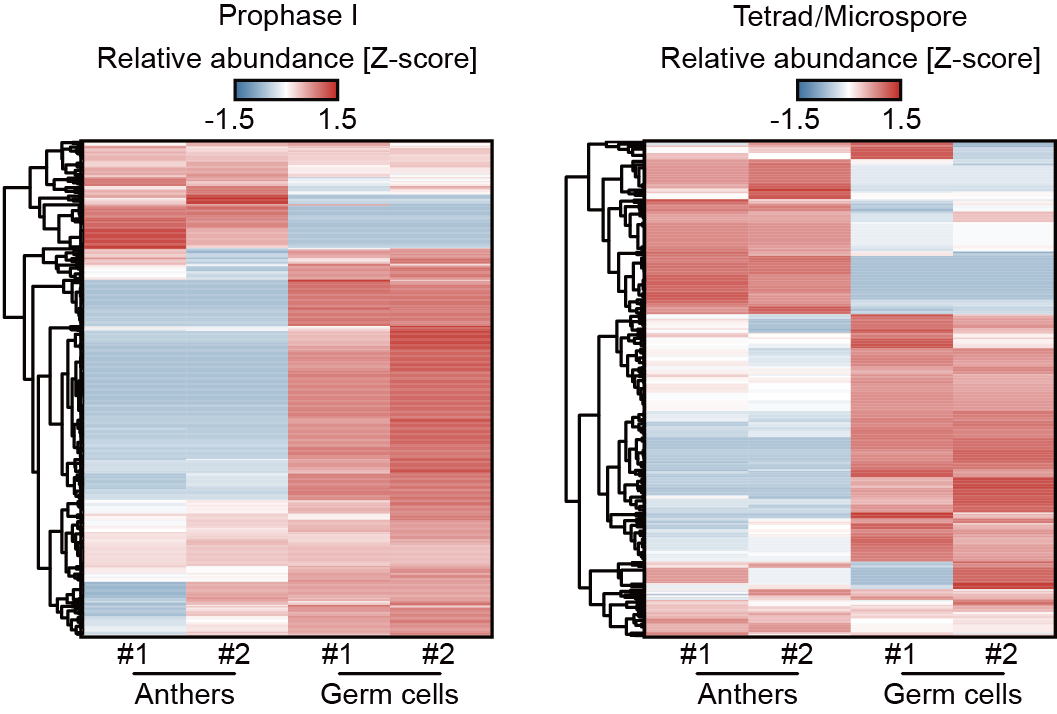
Fig. 3 Twenty-one-nt phasiRNA-directed cleavage signals in anthers and male germ cells.
Finally, evolutionary analysis revealed that 21-nt phasiRNAs are produced from conserved genomic locations in rice and maize. However, their sequences are entirely different, and they target distinct genes. These results suggest that 21-nt phasiRNA-producing loci and 21-nt phasiRNA targets have undergone fast divergence in grasses.
In summary, this study systematically identified target mRNAs that are cleaved by 21-nt phasiRNAs in maize anthers and male germ cells across various developmental stages, which provides a foundation for future investigation of phasiRNA biology in grasses. However, it remains elusive whether 21-nt phasiRNAs act collectively as a group to regulate male fertility or a few elite 21-nt phasiRNAs are responsible for the regulation.
The study, titled “Spatiotemporal regulation of target mRNA cleavage by 21-nt phasiRNAs in maize anthers”, was published in PNAS on June 17.
The study was co-corresponded by Professor Yijun Qi from the School of Life Sciences at Tsinghua University and postdoctoral researcher Bi Lian. Bi Lian and Tianxiao Huang, a 2019 Ph.D. candidate, served as co-first authors of the paper. Contributions were also made by Professor Pengfei Jiang from the School of Life Sciences at Anhui Agricultural University, as well as Dr. Wei Huang and Professor Weiwei Jin from the College of Agronomy and Biotechnology at China Agricultural University. This research was supported by fundings from the National Natural Science Foundation of China and the New Cornerstone Science Foundation.
Editor: Li Han

