An international team of scientists at Tsinghua and Peking university has developed a new computational method that can reconstruct the spatial organization of cells from single cell transcriptomic data. Published in Advanced Science, the study demonstrates that this method, termed De Novo Coalescent Embedding (D-CE), outperforms existing techniques in mapping single cells to their original locations within a tissue. This research is co-directed by Professor Carlo Vittorio Cannistraci, chief of the Center for Complex Network Intelligence (CCNI) at the Tsinghua Laboratory of Brain and Intelligence.
Single cell RNA sequencing (scRNA-seq) technologies profile gene expression in individual cells but lose information about the spatial context of the cells. Computational methods are needed to reconstruct the spatial relationships between cells based on their transcriptomic profiles. D-CE introduces an innovative network-based strategy to achieve this spatial mapping de novo, without requiring prior knowledge.
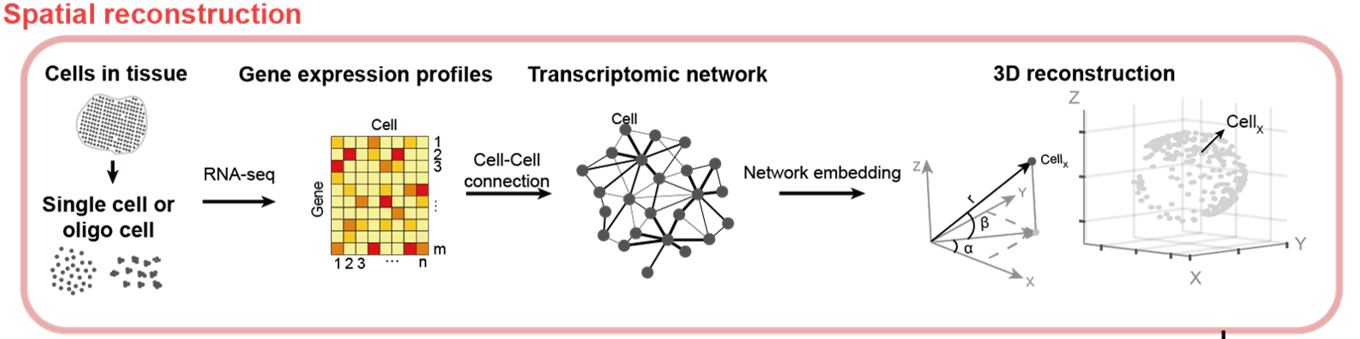 Figure 1. The new algorithm called de novo coalescent embedding (D-CE) for genome-based reconstruction of single cell 3D spatial tissue organization shows that it is possible to infer 3D spatial organization of single cells directly from their gene expression, by building a network of their transcriptomic profiles and performing the mapping of this network in a 3D geometrical space using coalescent embedding techniques.
Figure 1. The new algorithm called de novo coalescent embedding (D-CE) for genome-based reconstruction of single cell 3D spatial tissue organization shows that it is possible to infer 3D spatial organization of single cells directly from their gene expression, by building a network of their transcriptomic profiles and performing the mapping of this network in a 3D geometrical space using coalescent embedding techniques.
The researchers first tested D-CE on benchmark datasets with known spatial labels, showing it could accurately reconstruct cell locations in both mouse and human embryonic tissue samples. D-CE achieved superior performance compared to previous methods such as novoSpaRc and CSOmap, best separating cells from different spatial domains.
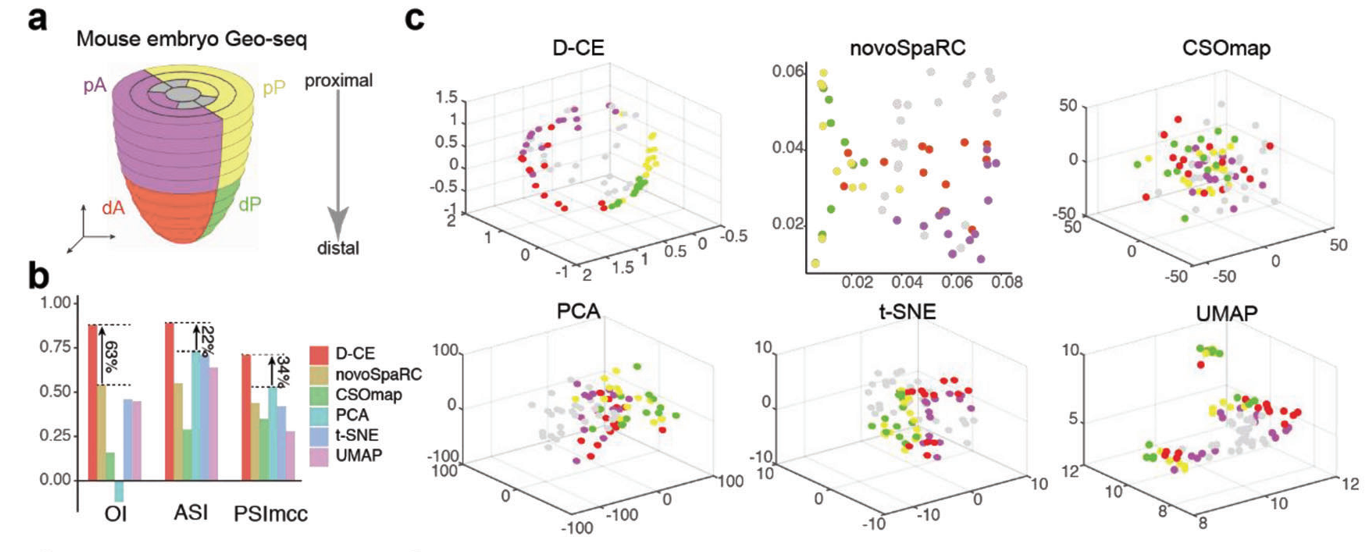
Figure 2. Reconstruction of spatial domain labels from oligo or single cell RNA-seq data with by D-CE and existing methods. a) Illustration of 4 spatial domains pA, dA, pP, and dP in mouse embryo E7.5. b) Barplot of OI, ASI, and PSImcc scores achieved by for D-CE, novoSpaRC, CSOmap, PCA, t-SNE, and UMAP reconstructions. These scores measures the accuracy of reconstruction, higher the better. c) Reconstructed structure of D-CE, novoSpaRC, CSOmap, PCA, t-SNE, and UMAP.
A key innovation of D-CE is its ability to also nominate marker genes expressed in specific spatial patterns, without prior knowledge. The researchers showed that using these D-CE-identified marker genes further enhanced spatial reconstruction accuracy.
Applying their approach to additional scRNA-seq datasets, the team demonstrated D-CE’s versatility in mapping spatial domains and cell distributions in fruit fly and zebrafish embryos. Overall, D-CE achieved at least 4-fold higher accuracy than existing methods across nearly 500 reconstructions of 14 datasets.
By reliably reconstructing spatial relationships between cells, D-CE revealed novel regulators of tissue patterning such as oxygen, matrix, and junction gradients. According to the researchers, D-CE’s accuracy and universality make it a valuable new tool for gaining biological insights from spatial patterns in single cell transcriptomic data.
Professor Jing-Dong J. Han from the Peking University School of Advanced Interdisciplinary Studies and Professor Carlo Vittorio Cannistraci from the Tsinghua University Brain and Intelligence Lab are co-corresponding authors of this paper. Ph.D. students Yuxuan Zhao and Shiqiang Zhang from Peking University Center for Quantitative Biology and Chinese Academy of Sciences Shanghai Institute of Nutrition and Health, respectively, are co-first authors. This research was funded by grants from the Ministry of Science and Technology of China, the National Natural Science Foundation of China (NSFC), and the Shanghai Municipal Science and Technology Project.
https://doi.org/10.1002/advs.202206307
Editor: Li Han

