Recently, Professor Zhucheng Chen’s group at the School of Life Science reported the crystal structure of human ALC1 (amplification in liver cancer 1,also known as CHD1L:chromodomain helicase/ATPase DNA binding protein 1-like) and the cryo-EM structure bound to the nucleosome. In combination with biochemical analyses, their findings shed light on the regulation mechanisms of the enzyme, paving the way for drug discovery targeting ALC1 for the treatment of cancer.
In eukaryotes, the genomic DNA is wrapped around histones to form nucleosomes, which are further assembled into chromatin. Packages of the chromatin within the nucleus block access to the DNA. Chromatin remodelers alter the positions and compositions of nucleosomes, regulating the chromatin structure and various nuclear transactions. Tight control of the activity of remodeling enzymes ensures the proper chromatin landscape and cellular functions. ALC1, a member of the Snf2 superfamily ATPases, contains two RecA-like ATPase domains (lobe 1 and lobe 2) and a C-terminal macro domain (Fig a). ALC1 relaxes chromatin and plays an important role in the poly (ADP-ribose) polymerase 1 (PARP1)-mediated DNA repair pathway. With DNA damage, PARP1 is activated and catalyzes the synthesis of poly (ADP-ribose) (PAR) chains on itself and on many other proteins. The PAR chains are bound to the macro domain of ALC1, releasing the autoinhibition of ALC1 and targeting the remodeler to the sites of DNA damage, where ALC1 slides the nucleosome and opens the chromatin.
ALC1 is amplified in over 50% of the cases of hepatocellular carcinoma (HCC). The oncogenic functions of ALC1 are widely demonstrated both in vitro and in vivo. A high level of ALC1 expression significantly reduces the postoperative survival rate of HCC patients. All of these indicate that ALC1 is an attractive target for cancer treatments. Despite its importance in DNA repair and clinical implication, the structure of ALC1 and its regulation mechanisms remain unclear.
It is a challenge to determine the structure of ALC1, as it is notoriously flexible. In 2017, two studies reported the possible structure models of ALC1, using the methods of hydrogen-deuterium exchange (HDX), mutagenesis, small-angle x-ray scattering (SAXS) and cross-linking mass-spectrometry (XL-MS). In order to stabilize the conformation of ALC1, Chen’s group screened a yeast-display library and obtained single chain antibodies (scFvs: single-chain antibody fragments/ single-chain variable fragments) directed against ALC1, which finally led to crystals diffracting to 3.5 Å (Fig b).
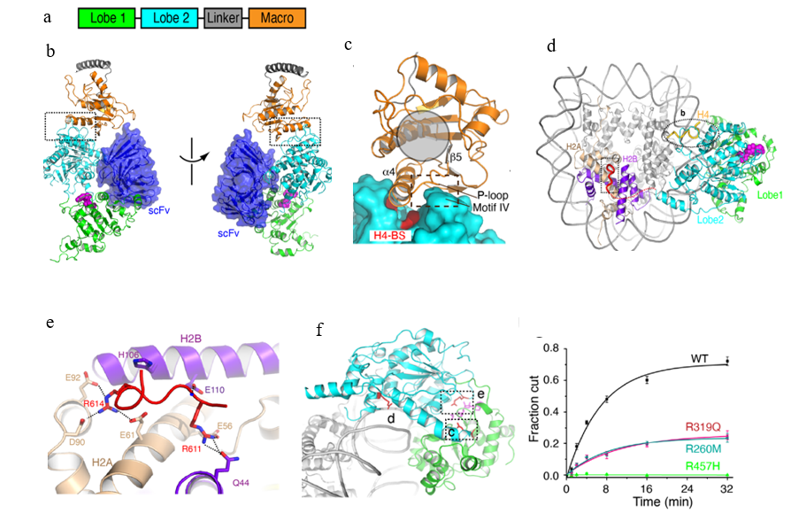
The crystal and the cryo-EM structures of ALC1
a. Domain architecture of ALC1
b. Two different views of the crystal structure of ALC1 in the autoinhibited state
c. The view of the macro-lobe 2 interaction
d. Top view of the cryoEM structure of the ALC1-nucleosome complex
e. Structure of the linker region of ALC1 bound to the acidic patch of the nucleosome
f. Dysregulation of ALC1 by mutations found in cancer patients
The structure shows that lobes 1 and 2 of ALC1 have little direct interaction, suggesting a large degree of conformational plasticity and explaining the difficulty of crystallizing ALC1 in the absence of the antibody. The macro-domain interacts with lobe 2 and sequesters two elements important for nucleosome recognition (Fig c), which provides the structural basis of ALC1 autoinhibition. To gain insights into ALC1 activation by the nucleosome, they determined the cryo-EM structure of ALC1 bound to the nucleosome (Fig d). Notably, in addition to the interactions with DNA and the H4 tail of the nucleosome, ALC1 is bound to the H2A-H2B acidic patch of the nucleosome (Fig e). The cancer-related mutations were found to dysregulate ALC1 by diverse mechanisms (Fig f). The structures and regulation mechanisms of ALC1 revealed in this study pave the way for the future discovery of drugs that target ALC1 in cancer treatments.
This work was published in “Nature Communications”, with the title of “Structural basis of ALC1/CHD1L autoinhibition and the mechanism of activation by the nucleosome”. It is an important part of the work by Chen's group on chromatin remodeling. Li Wang and Kangjing Chen, 2017 PhD students in the School of Life Science, Tsinghua University, are co-first authors of the paper and Professor Zhucheng Chen is the corresponding author. This work was supported by the National Natural Science Foundation of China, National Key Research and Development Program, Advanced Innovation Center for Structural Biology and Tsinghua-Peking Joint Center for Life Sciences. The Tsinghua University Branch of the China National Center for Protein Sciences Beijing provided facilities and the staff at beamline BL17U of Shanghai Synchrotron Radiation Facility helped with diffraction data collection.

