Zhucheng Chen Laboratory at the School of Life Science, Tsinghua University publishes a collaborative research article in Science
illustrating the mechanism of RSC in chromatin remodeling
On Nov. 1st, Zhucheng Chen (Tsinghua University), Ning Gao (Peking University), and Bradley Cairns (University of Utah, USA) published their collaborative research paper titled "Structure of the RSC complex bound to the nucleosome" online in the journal Science. This work analyzed the structure of the essential chromatin remodeling complex RSC bound to its substrate, the nucleosome, using cryo-electron microscopy (cryoEM), which reveals new information on the mechanism of chromatin remodeling mediated by the RSC complex.
Chromatin carries genetic information. It compresses DNA to suit the cellular space, but generally inhibits gene expression. Chromatin remodeler alters the position and composition of nucleosomes, which are the basic units of chromatin, playing an important role in enabling gene expression, DNA replication and DNA repair. RSC is a key chromatin remodeling complex and regulates expression of most of the genes in yeast, and is very similar to a complex in humans (termed PBAF) with similar roles. The RSC complex contains 15 subunits with a molecular weight above 1MDa. Although widely studied, how RSC is assembled, how it slides nucleosomes and opens up the promoter of genes has remained incompletely understood.
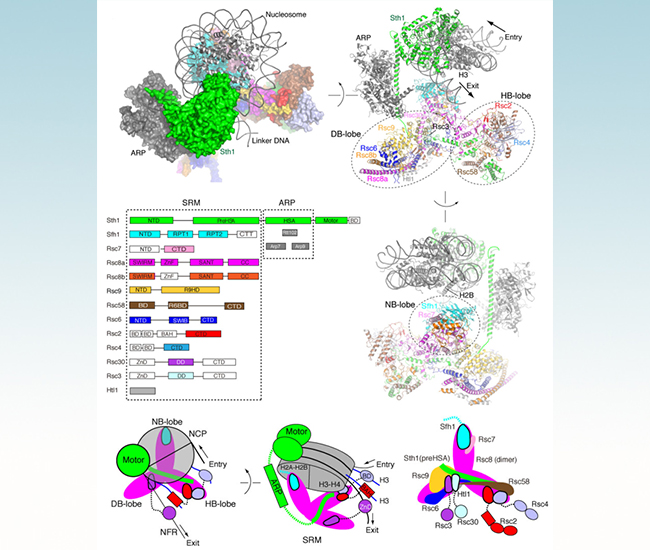
Figure 1 overall structure of RSC-nucleosome complex and the nucleosome recognition model
Zhucheng Chen lab has conducted a series of structural studies on the mechanism of chromatin remodeling (Yan, Nature 2016; Liu, Nature 2017; Li, Nature, 2019), that complement and extend biochemical studies from Bradley Cairns lab (Saha et al., Genes & Dev, 2002 and Nat. Str. & Mol. Biol., 2005). Based on these studies, Zhucheng Chen, in collaboration with Ning Gao and Bradley Cairns, determined the cryoEM structure of the RSC-nucleosome complex, and conducted in-depth functional studies. The structure shows that RSC is divided into three functional modules (Figure 1), namely, the motor module, the ARP module and the substrate recruitment module (SRM), which are organized through the core subunit Sth1. The motor module couples the energy of ATP hydrolysis to move the nucleosome. The ARP module mainly acts as a scaffold, and may influence the function of the ATPase. The SRM consists of three lobes: the DNA-binding lobe (DB-lobe), the histone binding lobe (HB-lobe) and the nucleosome binding lobe (NB-lobe). These lobes are poised near their corresponding substrates, enabling them to work independently and/or cooperatively to engage nucleosomes. This work also illustrates the mechanism of RSC in recognizing the promoter nucleosomes to open up the promoter of genes. Multiple subunits of RSC are highly conserved, and the structure of RSC provides an excellent template for understanding the human homologous complexes (PBAF).

Figure 2 Team photos
Youpi Ye (Ph.D student, Tsinghua University), and Hao Wu (Ph.D, Tsinghua university) are the co-first authors. Kangjing Chen (Ph.D student in Chen lab), and Wenhao Zhang (Ph.D student in Haiteng Deng’s lab) are two of the co-authors (Figure 2). Zhucheng Chen, Ning Gao, and Bradley Cairns are co-corresponding authors of this paper. This project is funded by the Ministry of Science and Technology of China, the National Natural Science Foundation of China(NSFC), and the Joint Center for Life Sciences of Tsinghua University and Peking University(CLS), and the Innovation Center for Advanced Structural Biology in Beijing(ICSB).
Related Link: https://science.sciencemag.org/content/early/2019/10/30/science.aay0033

